Group Profiles
Google Scholar Profile:
Xiao Wang
GitHub Page:
Wang Lab
Twitter Account:
WangXiaoLab
Addgene Account:
Xiao Wang Lab
Publications
*co-first authors; #corresponding authors
2024
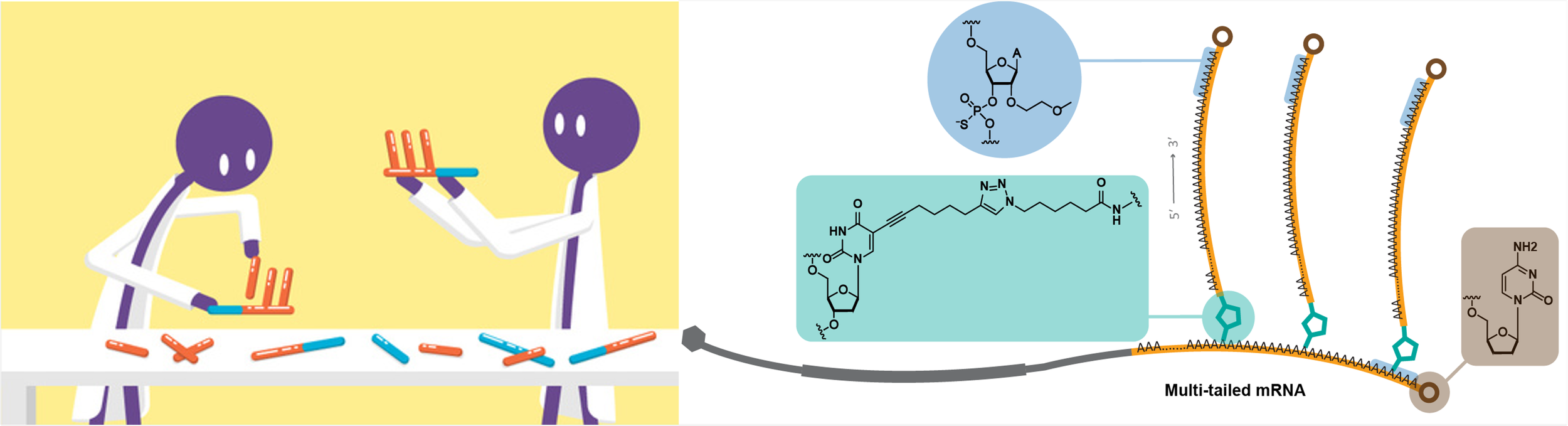
Hongyu Chen, Dangliang Liu, Jianting Guo, Abhishek Aditham, Yiming Zhou, Jiakun Tian, Shuchen Luo, Jingyi Ren, Alvin Hsu, Jiahao Huang, Franklin Kostas, Mingrui Wu, David R. Liu & Xiao Wang #. Branched chemically modified poly(A) tails enhance the translation capacity of mRNA. Nature Biotechnology 2024, https://doi.org/10.1038/s41587-024-02174-7 [Full text] [Broad News]
Preprints
Zefang Tang*, Shuchen Luo*, Hu Zeng, Jiahao Huang, Morgan Wu, Xiao Wang#. Search and Match across Spatial Omics Samples at Single-cell Resolution. bioRxiv 2023. [bioRxiv] [PDF]
Kamal Maher, Morgan Wu, Yiming Zhou, Jiahao Huang, Qiangge Zhang, Xiao Wang#. Mitigating autocorrelation during spatially resolved transcriptomics data analysis. bioRxiv 2023. [bioRxiv] [PDF]
2023
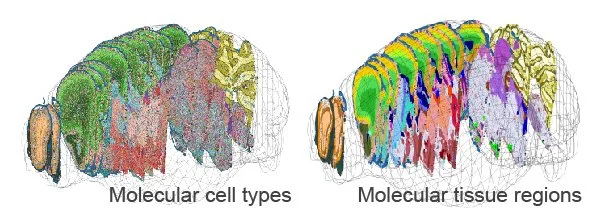
Hailing Shi*, Yichun He*, Yiming Zhou*, Jiahao Huang, Kamal Maher, Brandon Wang, Zefang Tang, Shuchen Luo, Peng Tan, Morgan Wu, Zuwan Lin, Jingyi Ren, Yaman Thapa, Xin Tang, Ken Y. Chan, Ben Deverman, Hao Shen, Albert Liu, Jia Liu#, Xiao Wang#. Spatial Atlas of the Mouse Central Nervous System at Molecular Resolution. Nature 2023, 622, 552-561. [Full text] [Data portal] [Code] [Plasmids] [bioRxiv 2022]
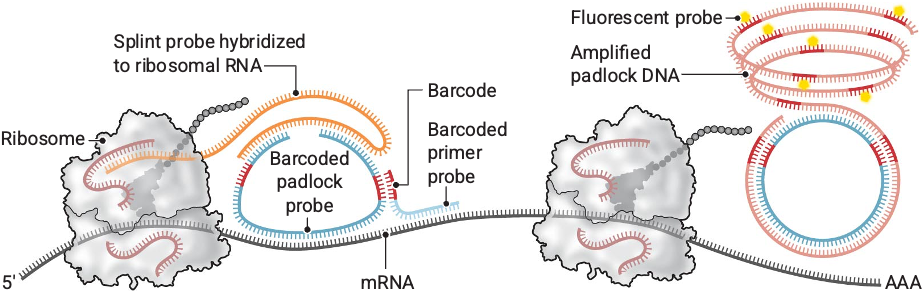
Hu Zeng*, Jiahao Huang*, Jingyi Ren*, Connie Kangni Wang, Zefang Tang, Haowen Zhou, Yiming Zhou, Abhishek Aditham, Hailing Shi, Xin Sui, Hongyu Chen, Jennifer A. Lo, Xiao Wang#. Spatially Resolved Single-cell Translatomics at Molecular Resolution. Science 2023, 380, eadd3067. [Full text] [Preprocessed Data] [Gene expression Data] [Code] [bioRxiv 2022] [Broad News] [Science Perspective]
Jingyi Ren*, Haowen Zhou*, Hu Zeng*, Connie Kangni Wang, Jiahao Huang, Xiaojie Qiu, Xin Sui, Qiang Li, Xunwei Wu, Zuwan Lin, Jennifer A. Lo, Kamal Maher, Yichun He, Xin Tang, Judson Lam, Hongyu Chen, Brian Li, David E. Fisher, Jia Liu, Xiao Wang#. Spatiotemporally resolved transcriptomics reveals the subcellular RNA kinetic landscape. Nature Methods 2023, 20, 695-705. [Full text] [PDF] [Data] [Code] [bioRxiv 2022] [Broad news]
Hu Zeng*, Jiahao Huang*, Haowen Zhou*, William J Meilandt, Borislav Dejanovic, Yiming Zhou, Christopher J Bohlen, Seung-Hye Lee, Jingyi Ren, Albert Liu, Zefang Tang, Hao Sheng, Jia Liu, Morgan Sheng#, Xiao Wang#. Integrative in situ mapping of single-cell transcriptional states and tissue histopathology in a mouse model of Alzheimer's disease. Nature Neuroscience 2023, 26, 430-446. [Full text] [PDF] [Data] [Code] [bioRxiv 2022] [Broad/MIT news] [Alzforum news/comments] [Research briefing]
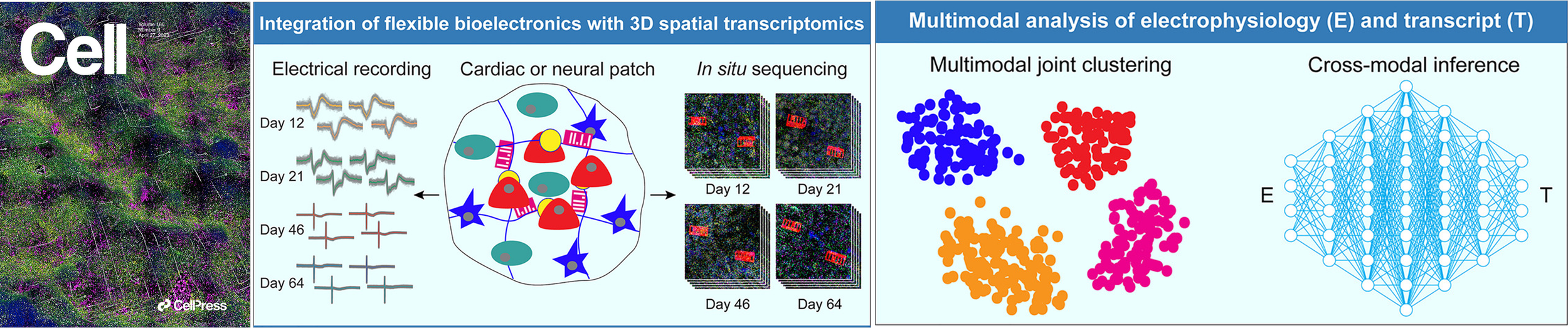
Qiang Li*, Zuwan Lin*, Ren Liu*, Xin Tang*, Jiahao Huang, Yichun He, Xin Sui, Weiwen Tian, Haowen Zhou, Hao Sheng, Hailing Shi, Ling Xiao, Xiao Wang#, Jia Liu#. Multimodal charting of molecular and functional cell states via in situ electro-sequencing. Cell 2023, 186, 2002-2017.e21. [Full text] [Data] [Code] [Cover art] [bioRxiv 2021]
Giridhar M Anand, Heitor C Megale, Sean H Murphy, Theresa Weis, Zuwan Lin, Yichun He, Xiao Wang, Jia Liu, Sharad Ramanathan. Controlling organoid symmetry breaking uncovers an excitable system underlying human axial elongation. Cell 2023, 186, 497-512.e23. [Full text]
Xin Tang, Jiawei Zhang, Yichun He, Xinhe Zhang, Zuwan Lin, Sebastian Partarrieu, Emma Bou Hanna, Zhaolin Ren, Hao Shen, Yuhong Yang, Xiao Wang, Na Li, Jie Ding & Jia Liu. Explainable multi-task learning for multi-modality biological data analysis. Nature Communications 2023, 14, 2546. [Full text]
2022
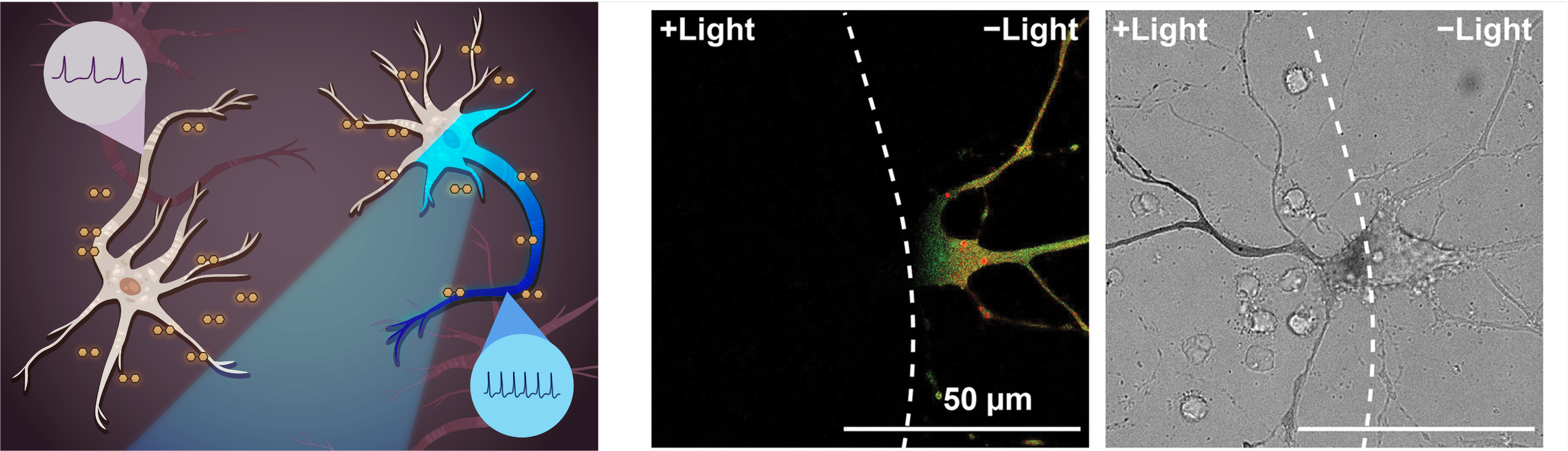
Chanan D Sessler*, Yiming Zhou*, Wenbo Wang*, Nolan D Hartley, Zhanyan Fu, David Graykowski, Morgan Sheng, Xiao Wang#, Jia Liu#. Optogenetic polymerization and assembly of electrically functional polymers for modulation of single-neuron excitability. Science Advances 2022, 8, eade1136. [Full text] [PDF] [Supplementary Information] [Plasmids] [MIT News] [Harvard News]
Xinyi Zhang, Xiao Wang, GV Shivashankar, Caroline Uhler#. Graph-based autoencoder integrates spatial transcriptomics with chromatin images and identifies joint biomarkers for Alzheimer’s disease. Nature Communications 2022, 13, 1-17. [Full text] [Code] [Broad News]
Emily L Sylwestrak, YoungJu Jo, Sam Vesuna, Xiao Wang, Blake Holcomb, Rebecca H Tien, Doo Kyung Kim, Lief Fenno, Charu Ramakrishnan, William E Allen, Ritchie Chen, Krishna V Shenoy, David Sussillo, Karl Deisseroth. Cell-type-specific population dynamics of diverse reward computations. Cell 2022, 19, 3568-3587. [Full text]
Albert Liu, Xiao Wang#. The Pivotal Role of Chemical Modifications in mRNA Therapeutics. Frontiers in Cell and Developmental Biology 2022, 10, 901510. [Mini-review] [Full text]
2021
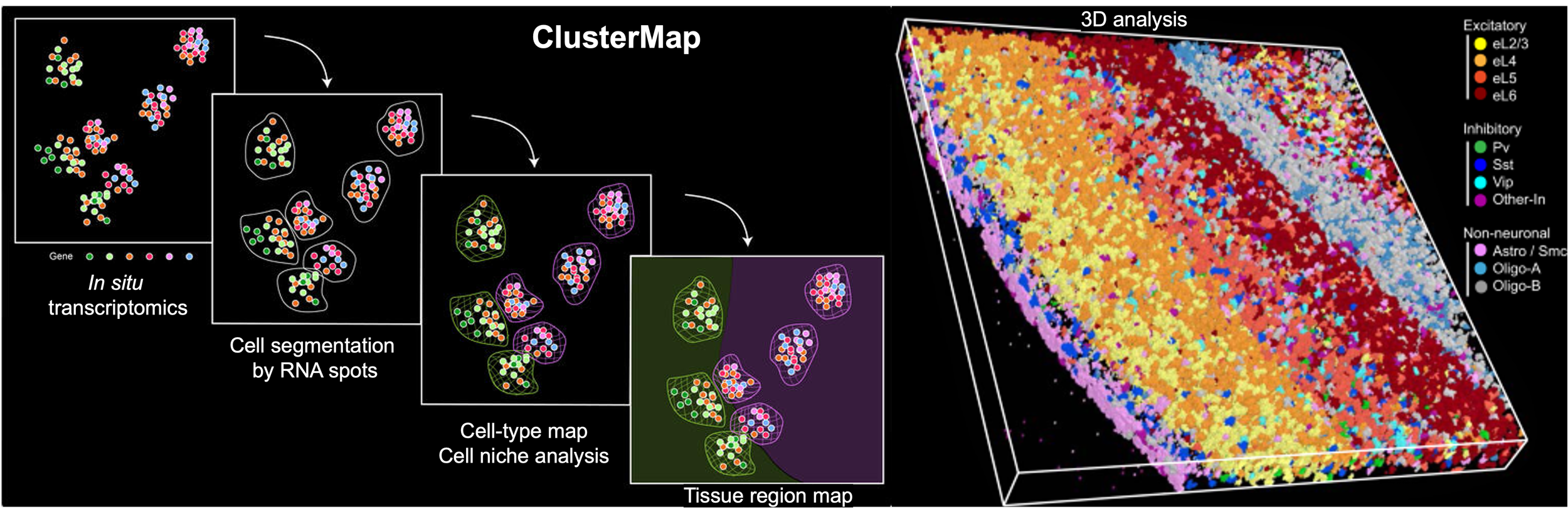
Yichun He*, Xin Tang*, Jiahao Huang, Jingyi Ren, Haowen Zhou, Kevin Chen, Albert Liu, Hailing Shi, Zuwan Lin, Qiang Li, Abhishek Aditham, Johain Ounadjela, Emanuelle I Grody, Jian Shu, Jia Liu#, Xiao Wang#. ClusterMap: multi-scale clustering analysis of spatial gene expression. Nature Communications 2021, 12, 5909. [Full text] [PDF] [Code] [Data]
Chanan D Sessler, Zhengkai Huang, Xiao Wang, Jia Liu#. Functional nanomaterial-enabled synthetic biology. Nano Futures 2021, 5, 022001. [Topical Review] [PDF]
Connor W Coley#, Xiao Wang. Editorial overview: Understanding, predicting, and optimizing biomolecular interactions with machine learning. Current Opinion in Chemical Biology 2021, 65, A1-A3. [Editorial overview] [Full text]
2020 and before
Selective publications:
X Wang*, W E Allen*, M Wright, E Sylwestrak, N Samusik, S Vesuna, K Evans, C Liu, C Ramakrishnan, J Liu, G P Nolan#, F-A Bava#, K Deisseroth#. Three-dimensional intact-tissue-sequencing of single-cell transcriptional states. Science 2018, eaat5691. [PDF] [Data + protocol]
B S Zhao*, X Wang*, A V Beadell*, Z Lu, H Shi, A Kuuspalu, R K Ho#, C He#. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017, 542, 475. [PDF]
X Wang*, B S Zhao*, I A Roundtree, Z Lu, D Han, H Ma, X Weng, K Chen, H Shi, C He#. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388. [PDF]
X Wang, Z Lu, A Gomez, G C Hon, Y Yue, D Han, Y Fu, M Parisien, Q Dai, G Jia, B Ren, T Pan, C He#. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117. [PDF]
Full list:
J Liu*, Y Kim*, C E Richardson*, A Tom*, C Ramakrishnan, F Birey, T Katsumata, S Chen, C Wang, X Wang, L-M Joubert, Y Jiang, H Wang, L E Fenno, J B-H Tok, S P Pașca, K Shen, Z Bao#, K Deisseroth#. Genetically targeted chemical assembly of functional materials in living cells, tissues, and animals. Science 2020, 367, 1372-1376. [PDF]
Y Liu*, J Liu*, S Chen, T Lei, Y Kim, S Niu, H Wang, X Wang, A M Foudeh, J B-H Tok, Z Bao#. Soft and elastic hydrogel-based microelectronics for localized low-voltage neuromodulation. Nature Biomedical Engineering 2019, 3, 58–68. [PDF]
X Wang*, W E Allen*, M Wright, E Sylwestrak, N Samusik, S Vesuna, K Evans, C Liu, C Ramakrishnan, J Liu, G P Nolan#, F-A Bava#, K Deisseroth#. Three-dimensional intact-tissue-sequencing of single-cell transcriptional states. Science 2018, eaat5691. [PDF]
I A Roundtree, G-Z Luo, Z Zhang, X Wang, T Zhou, Y Cui, J Sha, X Huang, L Guerrero, P Xie, E He, B Shen#, C He#. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. elife 2017,6, e31311. [PDF]
H Shi*, X Wang*, Z Lu, B S Zhao, H Ma, P J Hsu, C He#. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Research 2017, 27, 315. [PDF]
B S Zhao*, X Wang*, A V Beadell*, Z Lu, H Shi, A Kuuspalu, R K Ho#, C He#. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017, 542, 475. [PDF]
X Wang*, B S Zhao*, I A Roundtree, Z Lu, D Han, H Ma, X Weng, K Chen, H Shi, C He#. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388. [PDF]
K Chen, Z Lu, X Wang, Y Fu, G-Z Luo, N Liu, D Han, D Dominissini, Q Dai, T Pan, C He#. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angewandte Chemie International Edition 2015, 54, 1587. [PDF]
X Wang, Z Lu, A Gomez, G C Hon, Y Yue, D Han, Y Fu, M Parisien, Q Dai, G Jia, B Ren, T Pan, C He#. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117. [PDF]
T Zhu*, I A Roundtree*, P Wang, X Wang, L Wang, C Sun, Y Tian, J Li, C He, Y Xu#. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Research 2014, 24, 1493. [PDF]
X Wang, C He#. Dynamic RNA modifications in post-transcriptional regulation. Molecular Cell 2014, 56, 5. [perspective] [PDF]
C Xu*, X Wang*, K Liu*, I A Roundtree, W Tempel, Y Li, Z Lu, C He#, J Min#. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nature Chemical Biology 2014, 10, 927. [PDF]
X Wang, C He#. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biology 2014, 11. [point of view] [PDF]
J Liu*, Y Yue*, D Han, X Wang, Y Fu, L Zhang, G Jia, M Yu, Z Lu, X Deng, Q Dai, W Chen, C He#. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chemical Biology 2014, 10, 93. [PDF]
Y Fu, G Jia, X Pang, R Wang, X Wang, C Li, S Smemo, Q Dai, K Bailey, M Nobrega, K-L Han, Q Cui and C He#. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nature Communications 2013, 4, 1798. [PDF]
L Zhang, K E Szulwach, G C Hon, C-X Song, B Park, M Yu, X Lu, Q Dai, X Wang, C R Street, H Tan, J-H Min, B Ren, P Jin & C He#. Tet-mediated covalent labeling of 5-methylcytosine for its genome-wide detection and sequencing. Nature Communications 2013, 4, 1517. [PDF]
X Jiang, H Huang, Z Li, Y Li, X Wang, S Gurbuxani, P Chen, C He, D You, S Zhang, J Wang, S Arnovitz, A Elkahloun, C Price, G-M Hong, H Ren, R B Kunjamma, M B Neilly, J M Matthews, M Xu, R A Larson, M M Le Beau, R K Slany, P P Liu, J Lu, J Zhang, C He, J Chen#. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell 2012, 4, 524. [PDF]
X Wang, J Yan, Y Zhou, J Pei#. Surface modification of self-assembled 1D organic structures: white-light emission and beyond. Journal of the American Chemical Society 2010, 132, 15872. [PDF]
X Wang, Y Zhou, T Lei, N Hu, E-Q Chen, J Pei#. The structure-property relationship in pyrazino[2,3-g]quinoxaline derivatives: morphology, photophysical and waveguide properties. Chemistry of Materials 2010, 22, 3735. [PDF]
Resources
Spatial Mouse Brain Atlas:
mCNS database
Single cell portal
STARmap PLUS in Alzheimer’s brain:
Zenodo
Single cell portal
STARmap in prefrontal and visual cortices: STARmap Resources